Welcome to the community @jkozlowska.
Before I can answer your (many) questions, lets prepare the background and summarise all of the knowledge behind L2G that is currently implemented on the Platform.
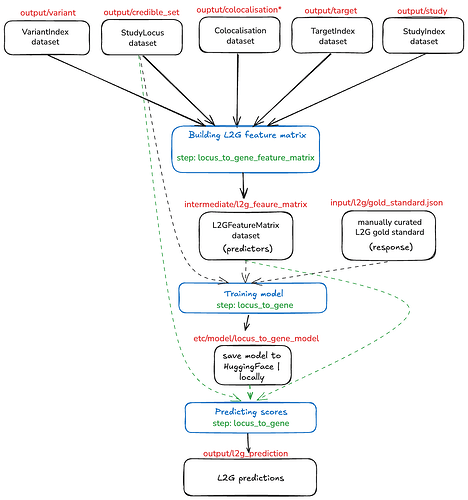
L2G steps breakdown
As you have found, to obtain the L2G scores we use gentropy steps. The whole process of obtaining the scores is divided into 3 parts:
- Building L2G Feature Matrix
- Training model
- Predicting scores
I had made a short schema of the process below
All dataset paths are written in red and are relative to the https://ftp.ebi.ac.uk/pub/databases/opentargets/platform/latest release.
This architecture is written in our [Unified Pipeline configuration](
Answers
Given the knowledge above, I can now answer your questions directly
- Can I use existing Open Targets L2G scores?
Yes, you can use the GraphQL api to retrieve scores for specific variant.
{
variant(variantId: "4_79957426_C_T") {
id
GWASCredibleSets: credibleSets(
studyTypes: [gwas]
) {
count
rows {
studyLocusId
studyId
l2GPredictions {
rows {
score
target {
id
}
}
}
}
}
}
}
This query should return the variant you were looking for and it’s L2G scores across all credible sets.
If you want to get the scores for loci associated with the disease of interest (assuming uc is Ulcerative Colitis ), you need to search for studies linked to the disease
{
studies(diseaseIds: "EFO_0000729") {
rows {
id
}
}
}
and finally filter results from the first query (all credible sets) by the studyIds from the second query. (not shown).
Note that the L2G score is not describing variant, rather full credible set. To have a best proxy of variant score estimation you would need to make sure that your variant has a high (>0.9) posterior inclusion probability within the searched credible set.
Is there a way to query/download L2G scores for my specific variant list? I see L2G data in the Open Targets Genetics portal, but I’m unclear on:
- Can I batch query with rsIDs or variant IDs?
- What’s the recommended approach: API, GraphQL, FTP download?
The preferred way to query in batch is to download the datasets
- predictions to find
studyLocusId and L2G scores
- credible_set to bring
variantId that account to studyLocusId
- study to filter out
studyLocusId by the relevant disease
Alternatively you can use big query to run SQL directly on the datasets.
- Do I need to/can I run L2G locally via gentropy?
After querying if you can not find your variants, to obtain L2G scores for them you would eventually need to run the fine-mapping (fortunately with in-sample-LD) and obtain credible sets.
To obtain the predictions for new credible sets You need to:
Building Feature Matrix
As you may have seen from the diagram above, to generate the L2G feature matrix, one need to provide:
- colocalisation dataset that contains results from colocalising your GWAS credible sets to QTL credible sets
- variant index that contains all variants from your GWAS credible sets annotated with VEP
- study index dataset that contains information about the molQTLs studies (the column
geneId is used to determine molecular feature affected by the credible set linked to study
- target index dataset - this can be used directly from the platform output without any modifications
Would L2G work on raw GWAS variants or is finemapping necessary for the pipeline to work?
No, the feature matrix step requires the PIP to be present for each individual variant in locus as it is used as a feature weight, so if you have variants, you need to run the fine-mapping before building L2G feature matrix.
- What’s the recommended workflow?
Given my starting point (GWAS variants + p-values only), what would you recommend:
Option A: Query existing Open Targets L2G scores
- Map my variants to OT study loci
- Download pre-computed L2G predictions
- Filter for my indications
Option B: Run fine-mapping first, then L2G
- Use SuSiE/FINEMAP to create credible sets
- Run gentropy L2G locally
- More rigorous but time-intensive
As mentioned above, I would start from querying existing predictions.
If you would not find the variants you are looking for, then I would suggest running fine-mapping and building the feature matrix, running L2G model with both..
By looking at your workflow ideas, I feel that you know most of the stuff already, if there is anything unclear, feel free to raise questions.
With kind regards,
Szymon Szyszkowski